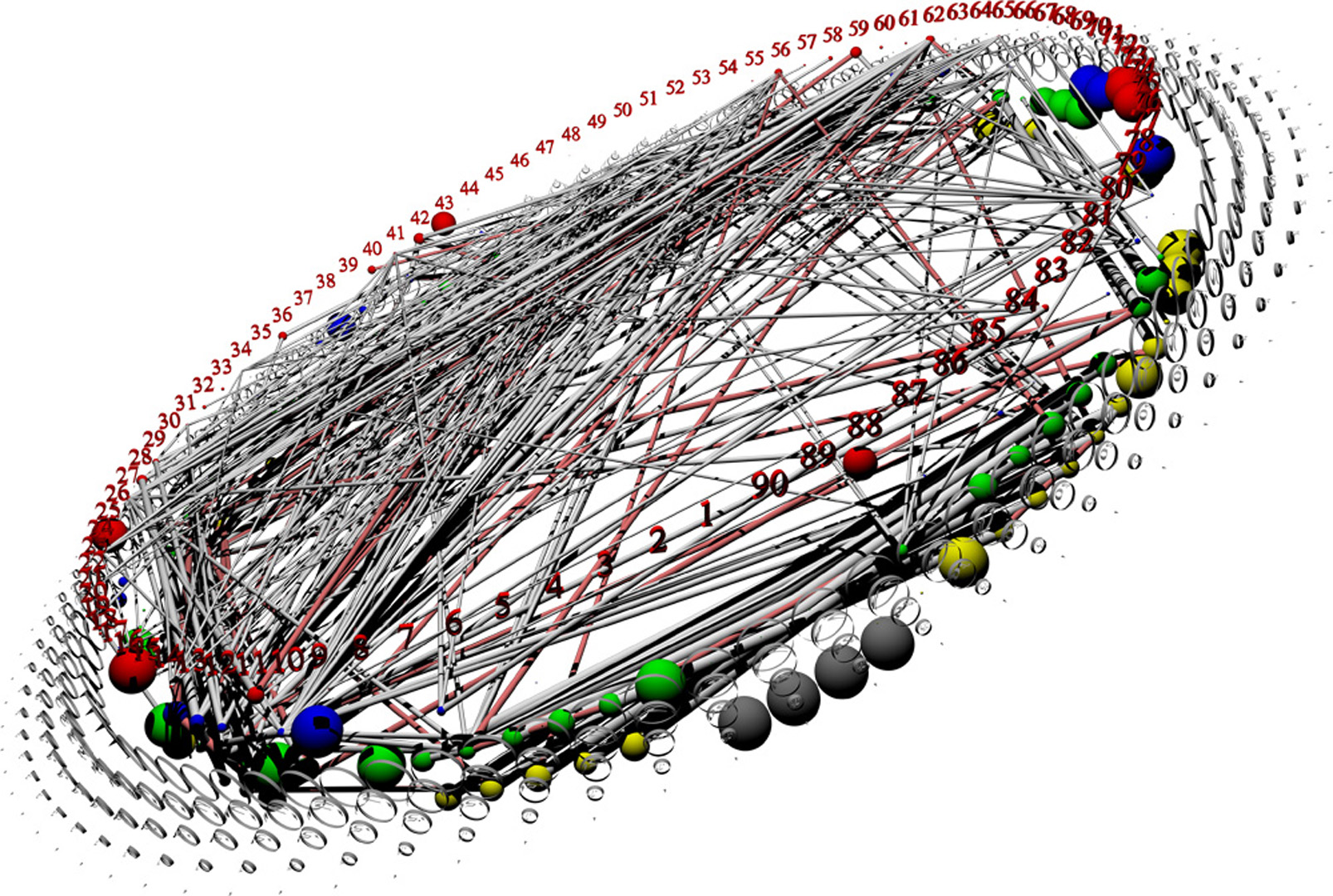
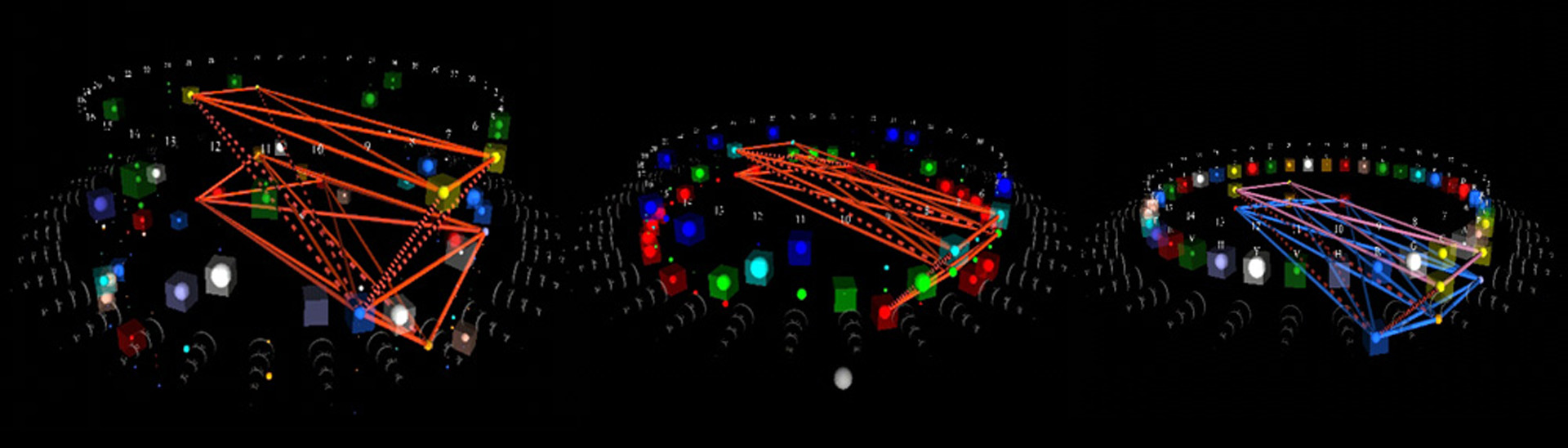
“Discovering biostructure constraints using VRML visualization” by Ray and Ozer
Conference:
Type(s):
Title:
- Discovering biostructure constraints using VRML visualization
Presenter(s)/Author(s):
Abstract:
The determination of a biomolecule’s complete 3-dimensional structure provides invaluable information about the molecule’s function, and how that function might be modulated. This in turn leads to insights across the biological spectrum. Unfortunately, determining protein or nucleic-acid structures from the molecules themselves is difficult due to physical impediments. Likewise, accurate computational prediction of such structures from physical principles remains computationally intractable due to the overwhelming number of degrees of freedom in the system.
References:
1. Berry, M., and Phillips, G. N. 1998. Crystal structures of Bacillus stearothermophilus adenylate kinase with bound Ap5A, Mg2+ Ap5A, and Mn2+ Ap5A reveal an intermediate lid position and six coordinate octahedral geometry for bound Mg2+ and Mn2+. Proteins: Structure, Function, and Genetics 32, 276–288.
2. Kawashima, S., and Kanehisa, M. 2000. AAindex: amino acid index database. Nucleic Acids Research 28, 374.