“Design and self-assembly of DNA into nanoscale 3D shapes” by Douglas, Dietz, Liedl, Högberg, Graf, et al. …
Conference:
Type(s):
Talk Type(s):
Title:
- Design and self-assembly of DNA into nanoscale 3D shapes
Session/Category Title:
- Rendering and Visualization
Presenter(s)/Author(s):
Abstract:
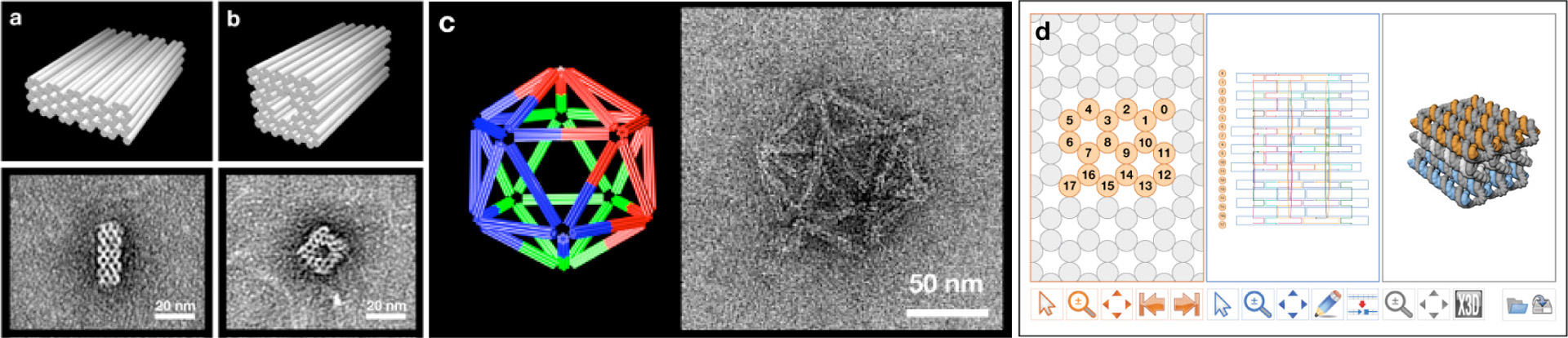
We have developed a general method for solving a key challenge for nanotechnology: programmable self-assembly of complex, three-dimensional nanostructures [Douglas, Dietz, et al. 2009]. Previously, scaffolded DNA origami has been used to build arbitrary flat shapes 100 nm in diameter and almost twice the mass of a ribosome [Rothemund 2006]. Now we have succeeded in building custom three-dimensional structures that can be conceived as stacks of nearly flat layers of DNA. Successful extension from two dimensions to three dimensions in this way depended critically on calibration of folding conditions. A general capability for building complex, three-dimensional nanostructures will be of great interest to biologists, chemists, physicists, engineers, and computer scientists.
References:
1. Dietz, H., Douglas, S. M., Shih, W. M. 2009. Folding DNA into twisted and curved nanoscale shapes. Science. In review.
2. Douglas, S. M., Dietz H., Liedl, T., Högberg, B., Graf, F., Shih, W. M. 2009. Self assembly of DNA into three-dimensional nanoscale shapes. Nature, 459, 414–418.
3. Douglas, S. M., Marblestone, A. M., Teerapittayanon, S., Vazquez, A., Church, G. M., Shih, W. M. 2009. Rapid prototyping of three-dimensional DNA-origami shapes with caDNAno. Nucleic Acids Res. In press.
4. Rothemund, P. W. K. 2006. Folding DNA to create nanoscale shapes and patterns. Nature, 440, 297–302.