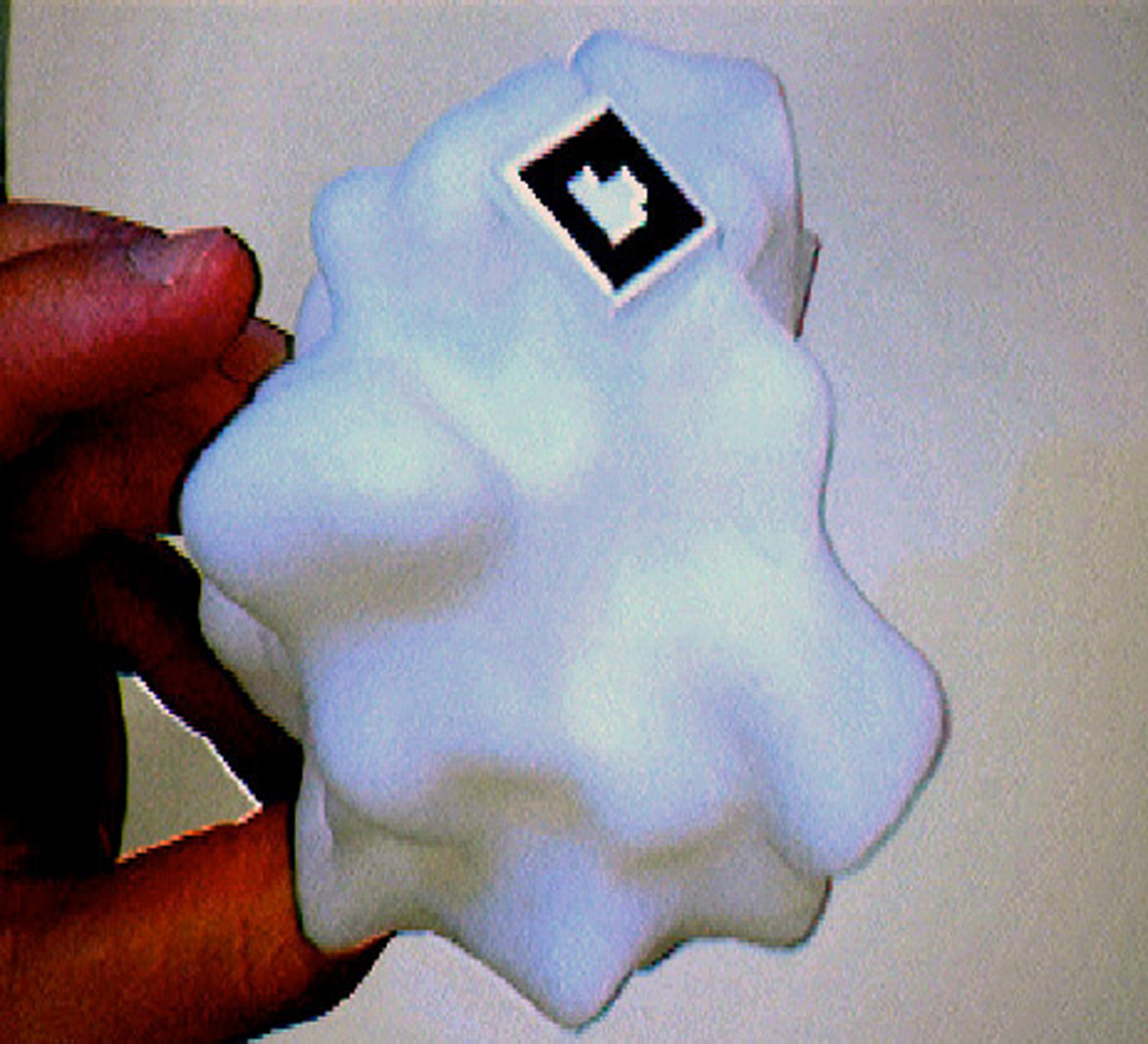
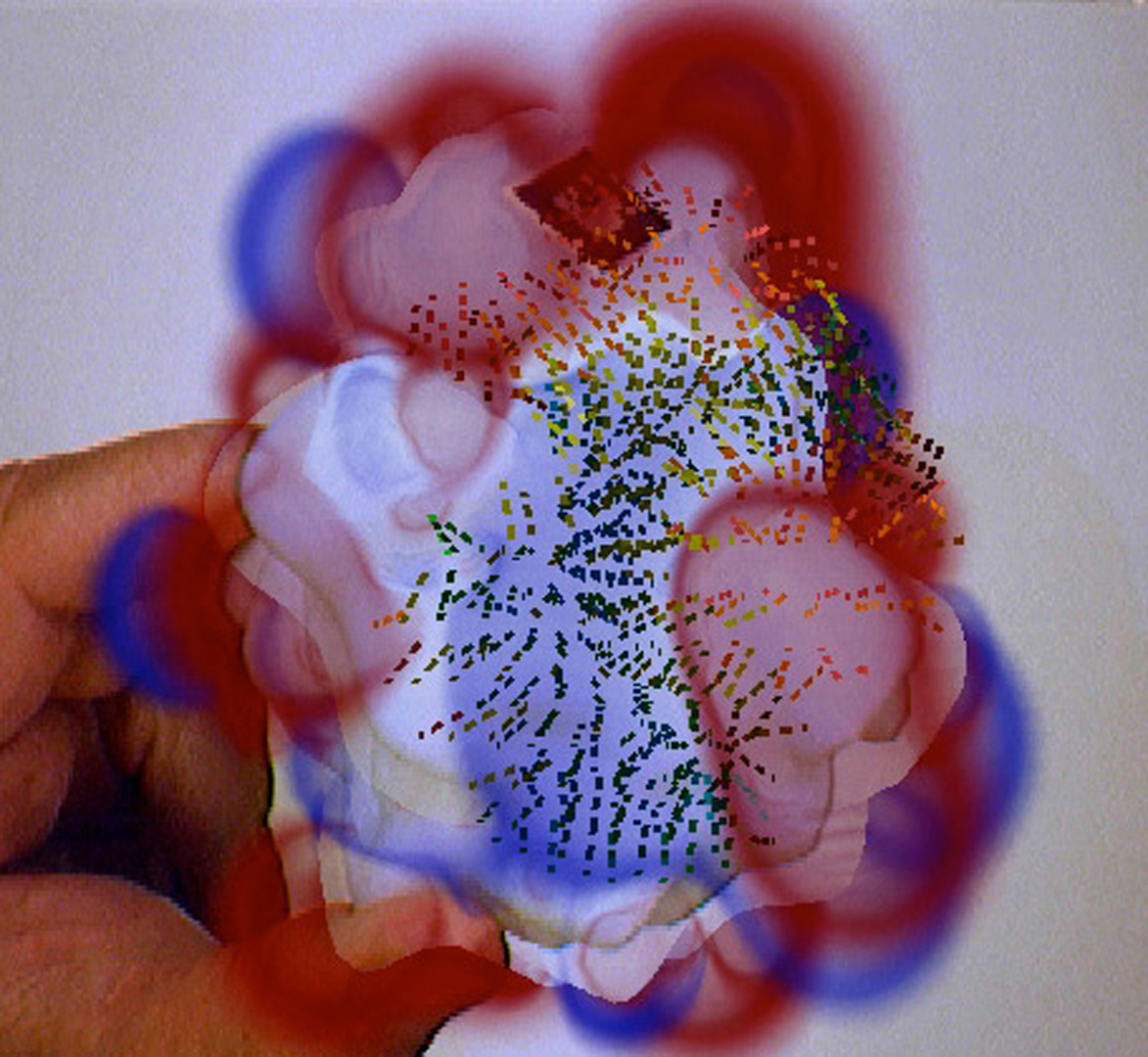
“Computer-linked autofabricated 3D models for teaching structural biology” by Olson, Gillet, Goodsell, Sanner, Stoffler, et al. …
Conference:
Type(s):
Title:
- Computer-linked autofabricated 3D models for teaching structural biology
Session/Category Title: Applications
Presenter(s)/Author(s):
Abstract:
research technologies – 3D printing and augmented reality – toward improving learning structural biology. Understanding this complex subject is essential in our society, both to foster progress and support critical decision- making in biotechnology and bio-nanotechnology. It is a challenging subject requiring the comprehension of the spatial structure and interactions among complex molecules comprising thousands of atoms. Our driving hypothesis is that the addition of augmented tangible elements to the perceptual experience of students will enhance and accelerate their understanding of structural molecular biology.
References:
Bailey, M., Schulten, K. and Johnson, J. (1998). “The use of solid physical models for the study of macromolecular assembly.” Curr Opin Struct Biol 8: 202–208.
Billinghurst, M. and Kato., H. (1999). “Collaborative mixed reality.” In Proceedings of International Symposium on Mixed Reality (ISMR ’99). Mixed Reality–Merging Real and Virtual Worlds: 261–284.
Sanner, M. F. (1999). “Python: a programming language for software integration and development.” J Mol Graph Model 17(1): 57–61