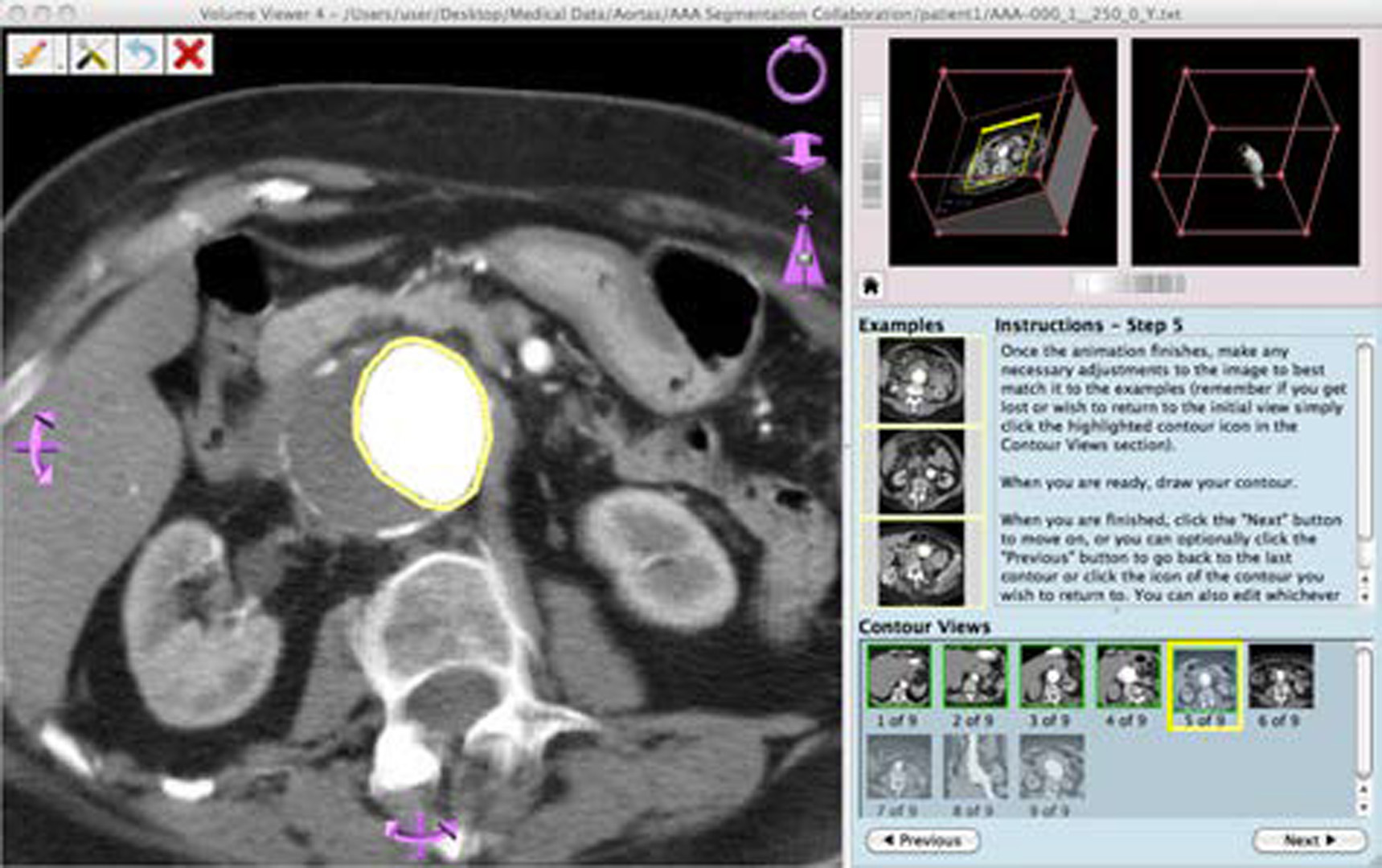
“A Guided Approach to Segmentation of Volumetric Data” by Holloway, Grimm, Sowell and West
Conference:
Type(s):
Entry Number: 93
Title:
- A Guided Approach to Segmentation of Volumetric Data
Presenter(s)/Author(s):
Abstract:
Recent advances in imaging techniques have made a fundamental impact on biomedicine. However, imaging produces volumetric data which, in its raw form, consists of colored pixels or voxels. This information needs to be processed in order to reveal useful knowledge about the physiology, morphology, and mechanics of the imaged subject. This data-to-knowledge pipeline usually starts with the delineation of the anatomical structure of interest (e.g., an organ, a tissue, a cell, etc.), a process commonly known as segmentation, to produce a 3D model of the structure(s).
Segmentations are critical for a wide range of processes found in practical applications such as: rendering visualizations; quantitative analysis of geometry, topology, shape, morphology, and statistics of image data within the structure; and computation of physical properties for treatment planning and virtual simulations. So not only do segmentations need to be accurate, but they also need to be created efficiently. When segmentation is the bottleneck in the data-to-knowledge pipeline, it slows the rate of research and analysis and can result in health risks in clinical practices [Foppiano et al. 2003], [Fiorino et al. 1998]. Automatic or semi-automatic segmentation methods are becoming more reliable [Wirjadi 2007], but in many cases they cannot be used because they require strong image gradients or templates. They are still highly susceptible to noise and poor image quality, thus requiring manual human interaction.
In this work, we consider reducing this significant bottleneck by creating a novice-friendly manual segmentation system that would allow crowdsourcing the task of segmenting 3D data. We focus on making volumetric data understandable and helping non-experts produce usable results. We use contouring guidelines, navigation aids, and other interface enhancements to allow any level of user to produce a 3D segmentation. This work builds off of [Sowell et al. 2009], a similar system designed for expert use.
References:
- Fiorino, C., Reni, M., Bolognesi, A., Cattaneo, G. M., and Calandrino, R. 1998. Intra- and inter-observer variability in contouring prostate and seminal vesicles: implications for conformal treatment planning. Radiother Oncol 47, 3, 285–92.
- Foppiano, F., Fiorino, C., Frezza, G., Greco, C., and Valdagni, R. 2003. The impact of contouring uncertainty on rectal 3D dose-volume data: results of a dummy run in a multicenter trial. Int J Radiat Oncol Biol Phys 57, 2, 573–9.
- Sowell, R., Liu, L., Ju, T., Grimm, C., Abraham, C., Gokhroo, G., and Low, D. 2009. VolumeViewer: an interactive tool for fitting surfaces to volume data. In SBIM ’09, ACM, New York, NY, USA, 141–148.
- Wirjadi, O. 2007. Survey of 3D image segmentation methods. Tech. Rep. 123, Fraunhofer-Institut fr Techno- und Wirtschaftsmathematik.
Acknowledgements:
Funded in part by NSF grants DEB 1053554 and IIS 1302142.