“Demand-driven volume rendering of terascale EM data” by Beyer, Hadwiger, Jeong, Pfister and Lichtman
Conference:
Title:
- Demand-driven volume rendering of terascale EM data
Session/Category Title: Volume and rendering
Presenter(s)/Author(s):
Abstract:
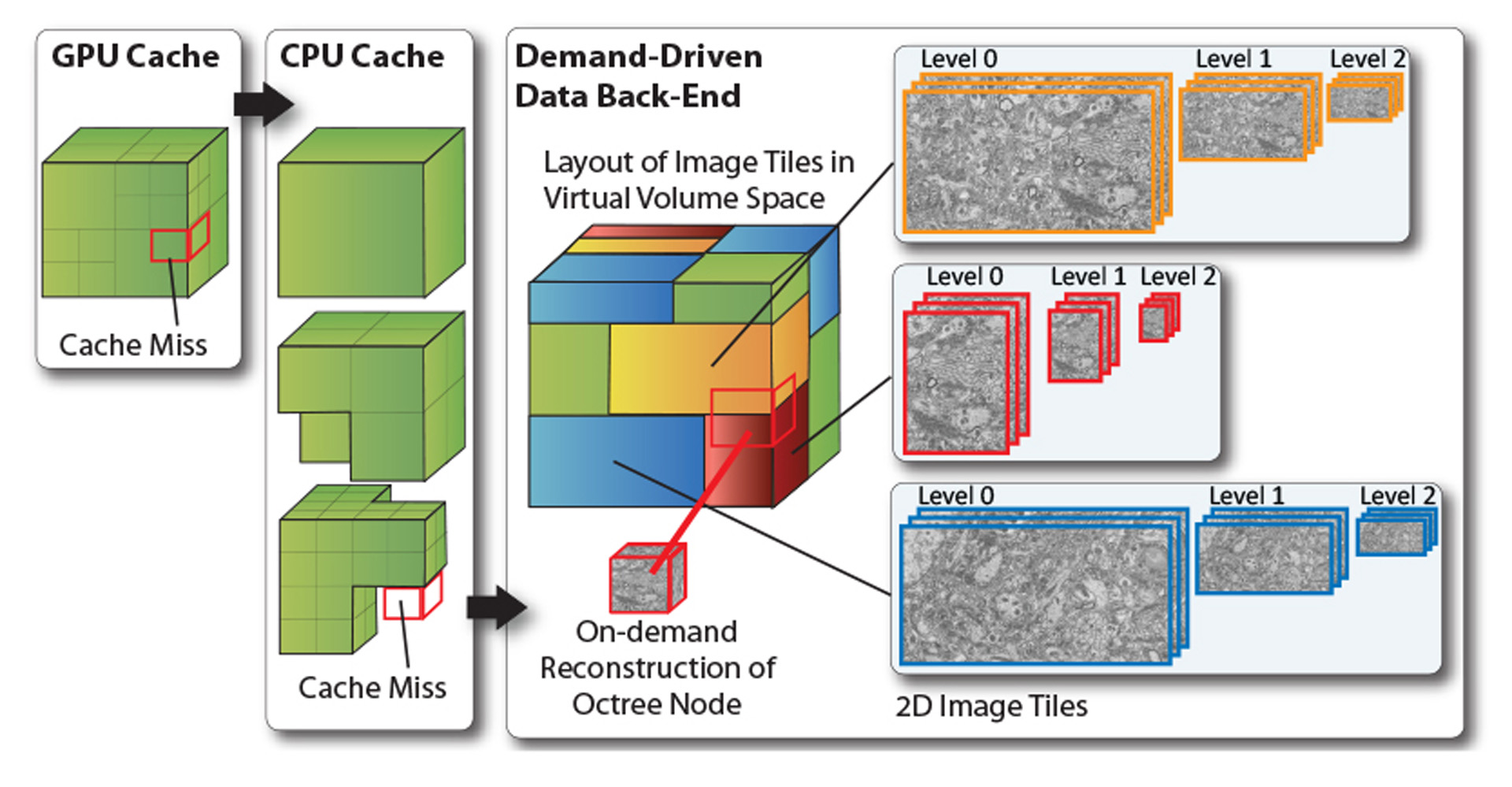
In neuroscience, a very promising bottom-up approach to understanding how the brain works is built on acquiring and analyzing electron microscopy (EM) scans of brain tissue, an area known as Connectomics. This results in volume data of extremely high resolution of 3–5nm per pixel and 25–50nm slice thickness, overall leading to data sizes of many terabytes [Jeong et al. 2010]. To support the work of neurobiologists, interactive exploration and analysis of such volumes requires novel visual computing systems because the requirements differ from those of current systems in several key aspects. In this talk, we describe the system that we are working on to enable neuroscientists to interactively roam terascale EM volumes and support their analysis. A major design principle was to avoid the standard approach of pre-computing a 3D multi-resolution hierarchy such as an octree. Data acquisition proceeds from 2D image tile to 2D image tile, where not only the slices along the z axis are scanned independently, but each slice is itself acquired as many smaller image tiles. These images tiles need to be aligned and stitched, and neurobiologists also want to be able to combine different resolutions used for scanning different regions, without re-sampling everything to a single global resolution. Therefore, we focus on working directly with a stream of individual 2D image tiles, instead of a 3D volume that usually is assumed to exist in its entirety for visualization. We perform interactive volume rendering of a “virtual” volume, where the corresponding physical storage is only represented and populated in a sparse manner with 2D instead of 3D image data on the fly during rendering. Furthermore, these 2D image tiles can be of different resolution, scale, and orientation.
References:
1. Crassin, C., Neyret, F., Lefebvre, S., and Eisemann, E. 2009. GigaVoxels: Ray-Guided Streaming for Efficient and Detailed Voxel Rendering. In Proceedings of 2009 Symposium on Interactive 3D Graphics and Games, 15–22.
2. Gobbetti, E., Marton, F., and Guitan, J. 2008. A Single-pass GPU Ray Casting Framework for Interactive Out-of-core Rendering of Massive Volumetric Datasets. The Visual Computer 24, 7, 797–806.
3. Jeong, W.-K., Beyer, J., Hadwiger, M., Blue, R., Law, C., Vasquez, A., Reid, C., Lichtman, J., and Pfister, H. 2010. SSECRETT and Neuro-Trace: Interactive Visualization and Analysis Tools for Large-Scale Neuroscience Datasets. IEEE Computer Graphics and Applications 30, 3, 58–70.